research
Our lab develops machine learning methods for high-dimensional, sparse, heterogeneous, and multimodal genomics data. Our work is motivated by the gap that while cellular and molecular systems are highly complex and dynamic, individual experimental data often capture partial and limited snapshots of biological processes. To address this challenge, we develop network-based and deep learning approaches that integrate large-scale public datasets, including both bulk and single-cell data, along with prior biological knowledge. Our research interests lies in the following areas:
- predicting multimodal representations of cells;
- transfering experimental insights from model organisms to human contexts;
- identifying genes and pathways underlying human diseases and sex differences.
multimodal machine learning for biology
Biological systems can be represented through diverse assay types, or data modalities, each capturing a unique layer of molecular information and representing a partial view of the underlying system. Although biological processes and disease progression are inherently dynamic and involve multiple molecular layers, most high-throughput measurements capture only static snapshots. Our lab develops deep learning models that integrate large-scale public omics datasets to reconstruct multimodal cellular states and infer dynamic relationships across data modalities.
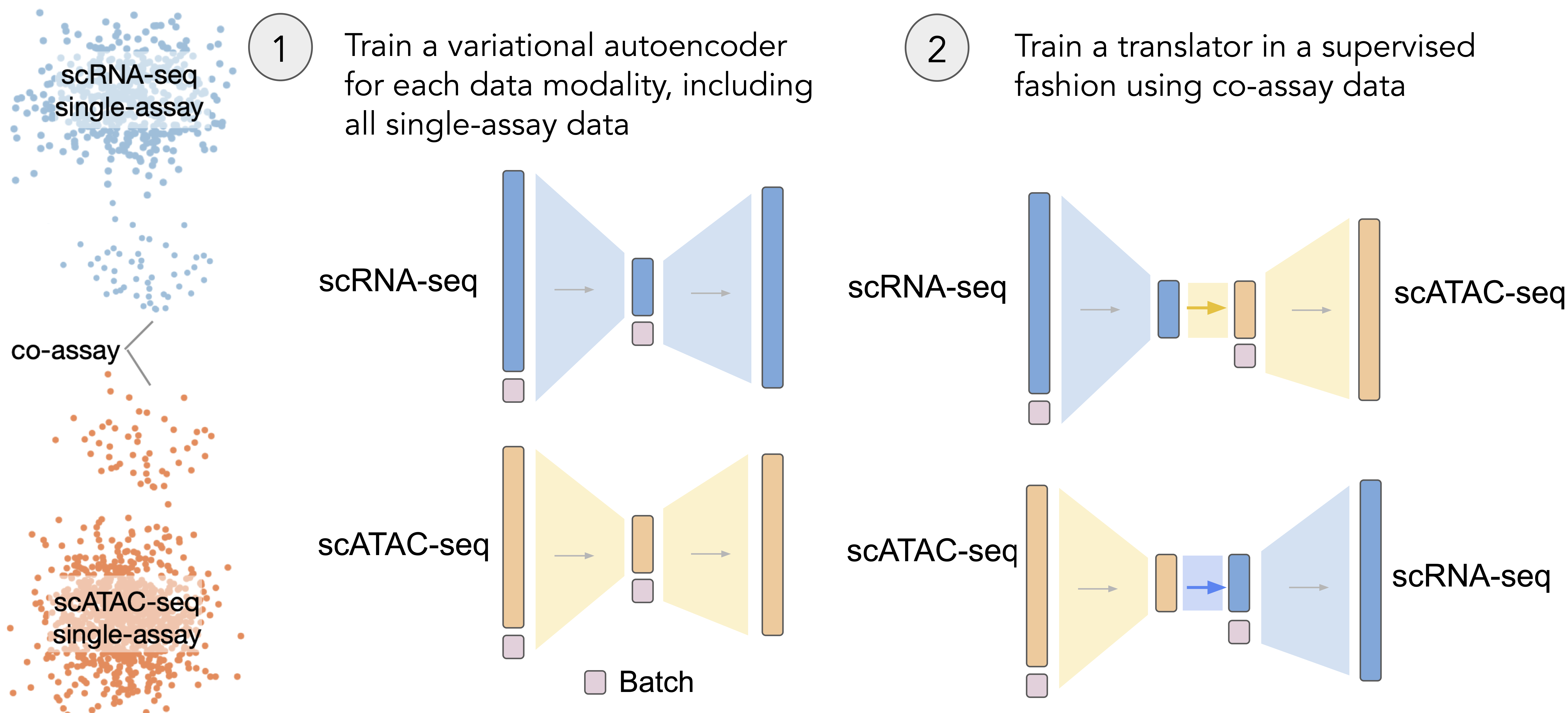
Selected publications:
- ”Semi-supervised single-cell cross-modality translation using Polarbear”
- ”Multimodal Single- Cell Translation and Alignment with Semi-Supervised Learning”
- ”Multicondition and multimodality single-cell temporal profile inference during mouse embryonic development”
network-guided discovery of human disease genes
Biological systems operate through complex networks of interacting molecules that vary across tissues and cell types. We design network-based models to integrate multimodal omics data, uncover molecular interactions, and identify genetic drivers of disease in their specific biological contexts.
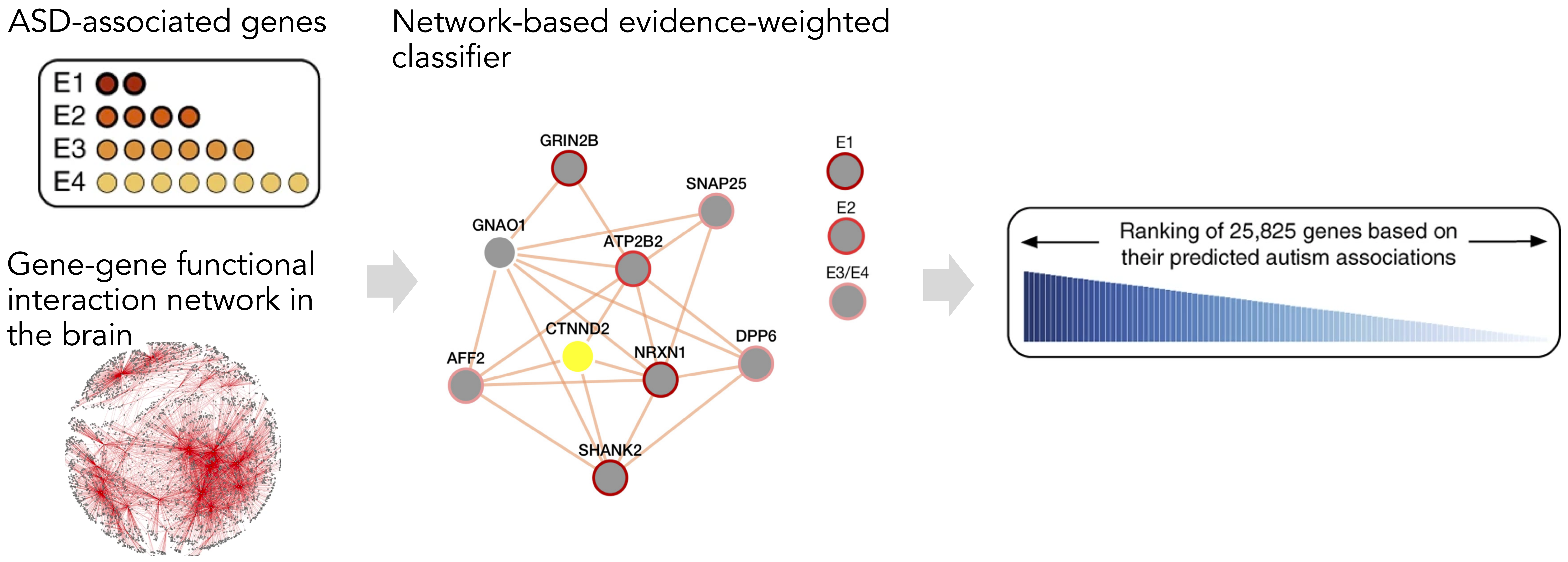
Selected publications:
- “Genome-wide autism gene prediction and functional characterization”
- “Lack of a site-specific phosphorylation of Presenilin 1 disrupts microglial gene networks and progenitors during development”
- “Astrocyte-derived extracellular matrix proteins regulate synapse remodeling in stress-induced depression”
knowledge transfer across species
Model organisms, such as mice, have been widely used to uncover molecular mechanisms relevant to human biology and diseases, especially when human samples are limited or inaccessible (e.g., the brain). However, evolutionary divergence between species create challenges for direct knowledge transfer, often limiting the translational value of findings. Our lab develops network and deep learning models to align, project, and contextualize data from model organisms, with the goal of improving data-driven insights into human disease.
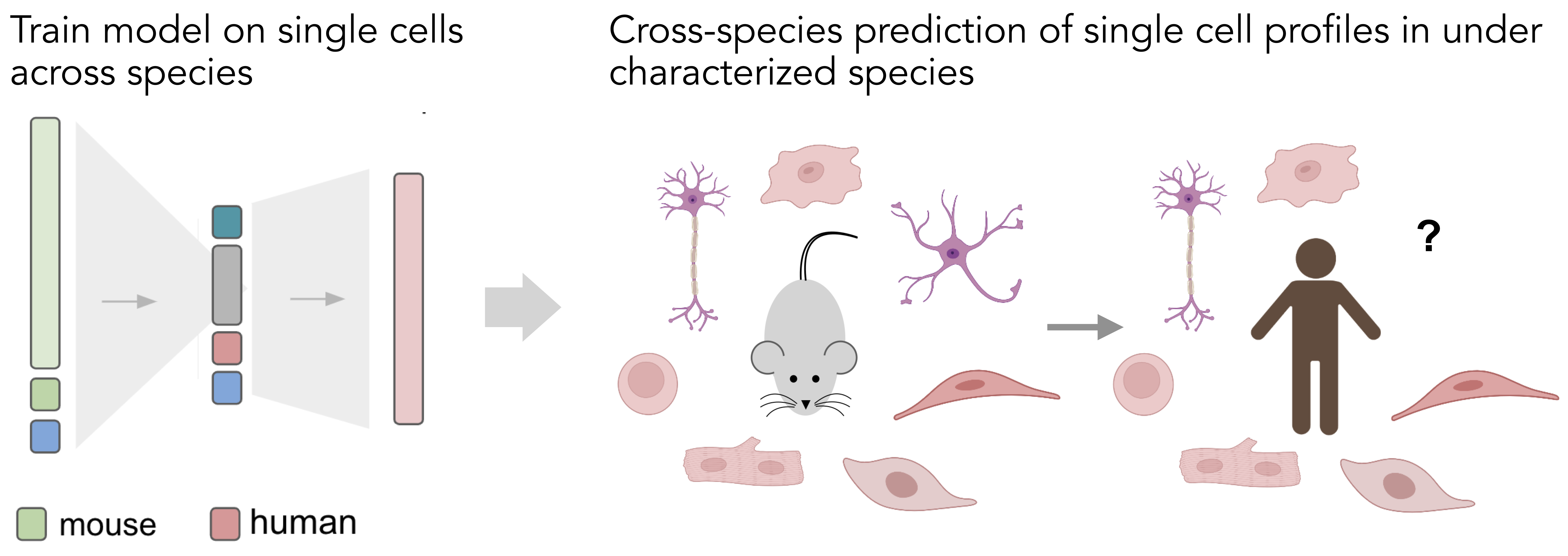
Selected publications: